As SARS-CoV-2 variants (mutated strains of the virus causing COVID-19) surface in different parts of the world, researchers rise to the challenge of examining tremendous amounts of new data and contextualizing it within what we already know about other strains.
Over the past year, researchers around the world have been dedicated to surveilling SARS-CoV-2 as the virus mutates. New variants are identified around the world every week. It is instrumental for researchers to ascertain whether new variants share mutations in common with existing variants known to have functional changes (i.e. changes in transmissibility, virulence, and/or antigenicity). Researchers also have an interest in tracking the growth and evolution of variants, so the world can begin to make plans for where the pandemic may be heading. With currently over 1,500 identified PANGO lineages and over 2 million sequenced SARS-CoV-2 samples, the amount of collected genomic data is enormous, and growing every day. Outbreak.info provides an easy way to get an overview of emerging lineages and examine changes in their genetic structure.
Researchers from the Robert Koch Institute (RKI) in Berlin use Outbreak.info to access genomic data and assess the prevalence of characteristic mutations in various lineages. Bioinformatician Dr. Martin Hölzer uses Outbreak.info daily to check the differences and similarities between lineages, sharing the resource he uses most regularly is the Compare Lineages tool. When a new lineage emerges, Hölzer and his colleagues can quickly go to Compare Lineages, add the lineage in question, and compare its mutations with known Variants of Concern/Interest (VOC/VOI). This gives them a preliminary idea of how concerning it might be, based on what is already known about the phenotypes of some Mutations of Concern/Interest (MOC/MOI).
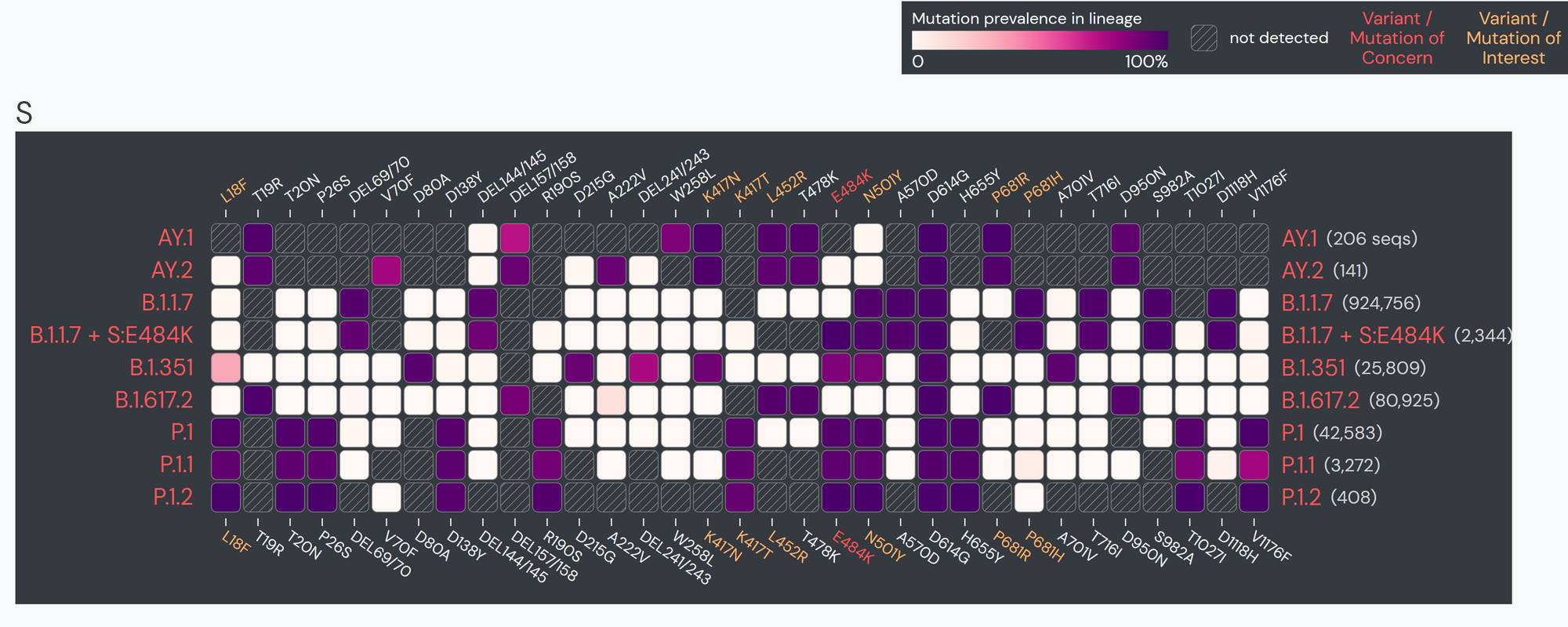
The research team at RKI frequently use the Lineage|Mutation Tracker to check lineage-specific reports when they need to access mutation profiles of specific lineages. The tool allows them to quickly find the characteristic mutations in the different genes. The tool also provides the ability to download the raw data and export figures in an easily reusable format.
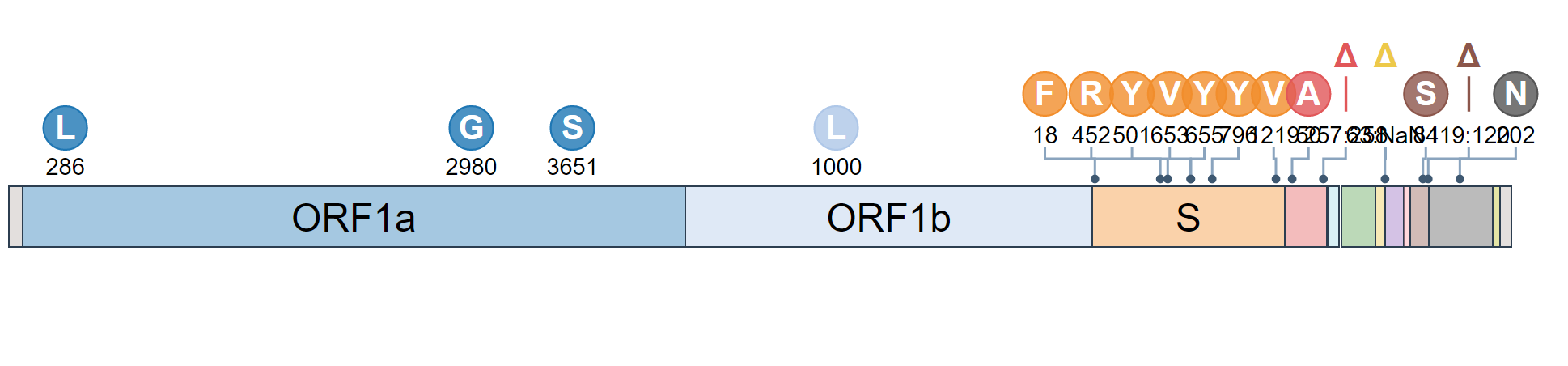
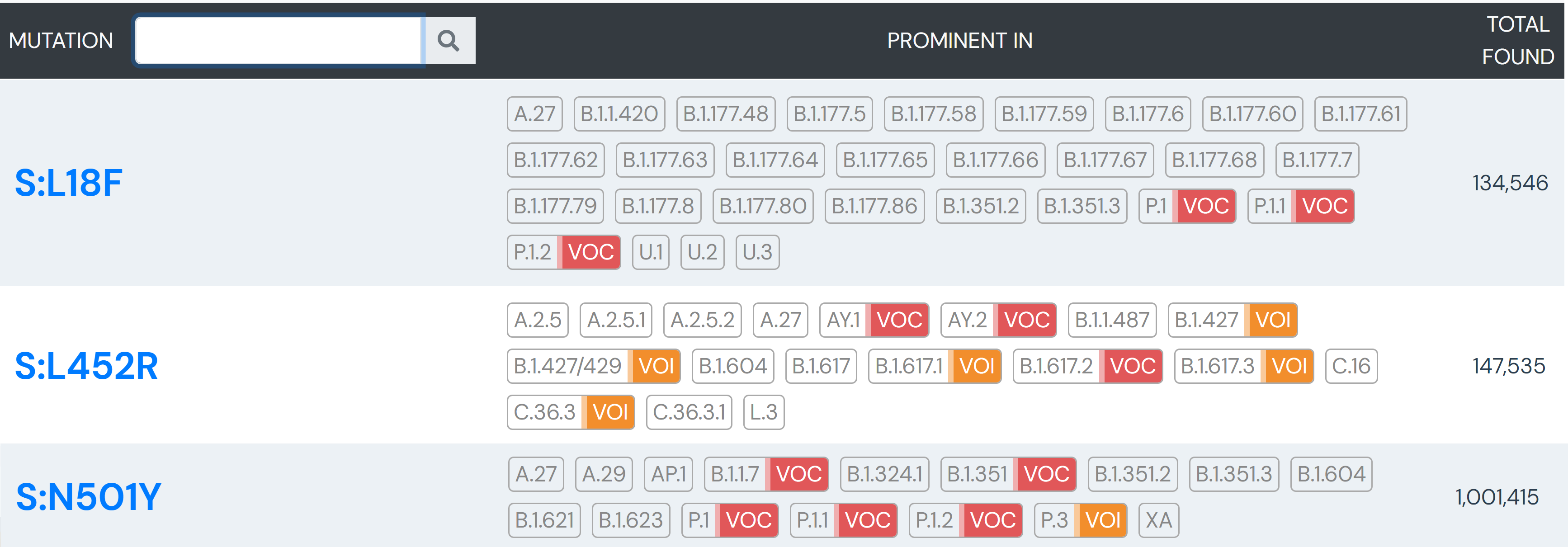
The team specifically used Outbreak.info’s research tools when investigating SARS-CoV-2 lineage A.27, upon sequencing the first samples in Germany in early 2021. A.27 is defined by 17 mutations, seven of which are nonsynonymous substitutions in the spike protein. Using the Compare Lineages tool, they discovered that some of these mutations are found in VOCs, including B.1.1.7 (Alpha variant originating in the United Kingdom), B.1.351 (Beta variant originating in South Africa), and P.1 (Gamma variant originating in Brazil). A.27 contains three worrying MOIs that may be associated with immune escape and reinfection. The S:L18F mutation (amino acid substitution of leucine to phenylalanine at position 18 in the S-gene; read more about mutations here) found in A.27 shows up in VOCs B.1.351 and P.1. Additionally, S:L452R appears in VOCs B.1.427/429, B.1.526.1, and B.1.617.2. This mutation shows evidence it may contribute to increased viral infectivity and enhanced viral replication. S:N501Y appears in VOCs B.1.1.7, P.1, and B.1.351, and shows some evidence of contributing to antibody resistance.
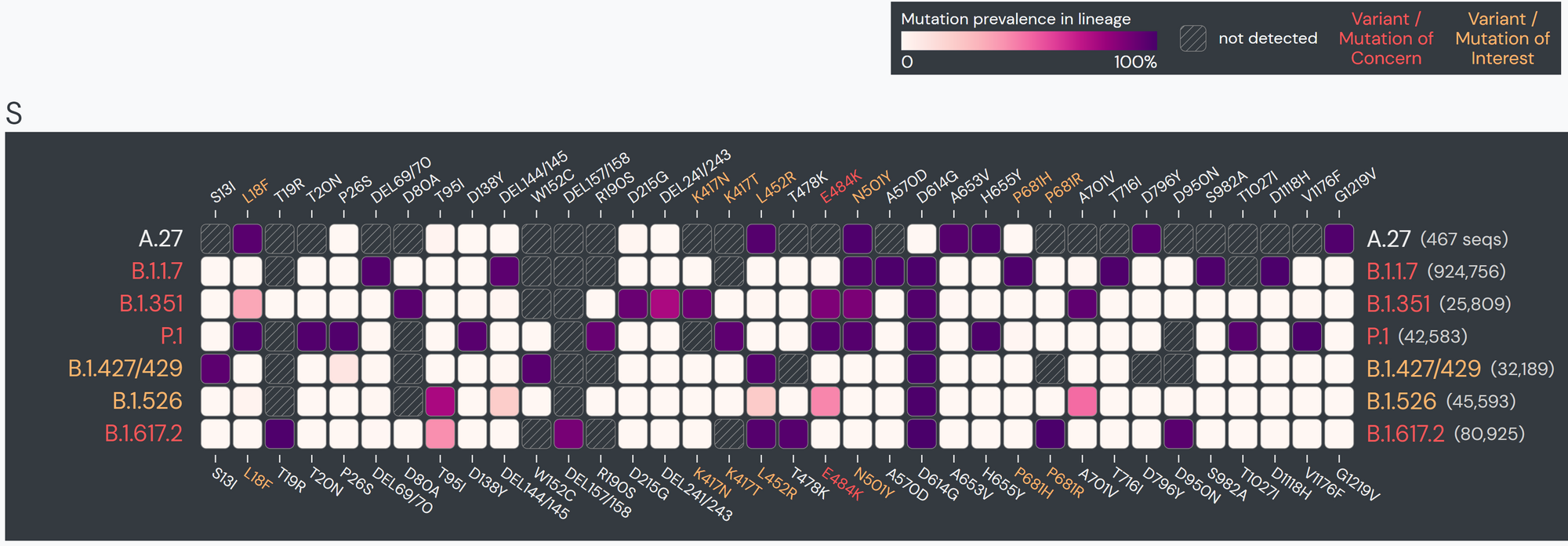
Beyond their search for the overlapping mutations, the team used Outbreak.info to find the differences between A.27 and other known variants. They found that A.27 generally does not possess mutation S:D614G, which is present in many dominant lineages around the world. A.27 does not contain mutation S:Q613H that is prevalent in A.23.1. The absence of these mutations provides clues about how A.27 may have separated from globally circulating variants early on. Moreover, it helps paint the picture of how certain mutations affect the fitness of SARS-CoV-2 variants.
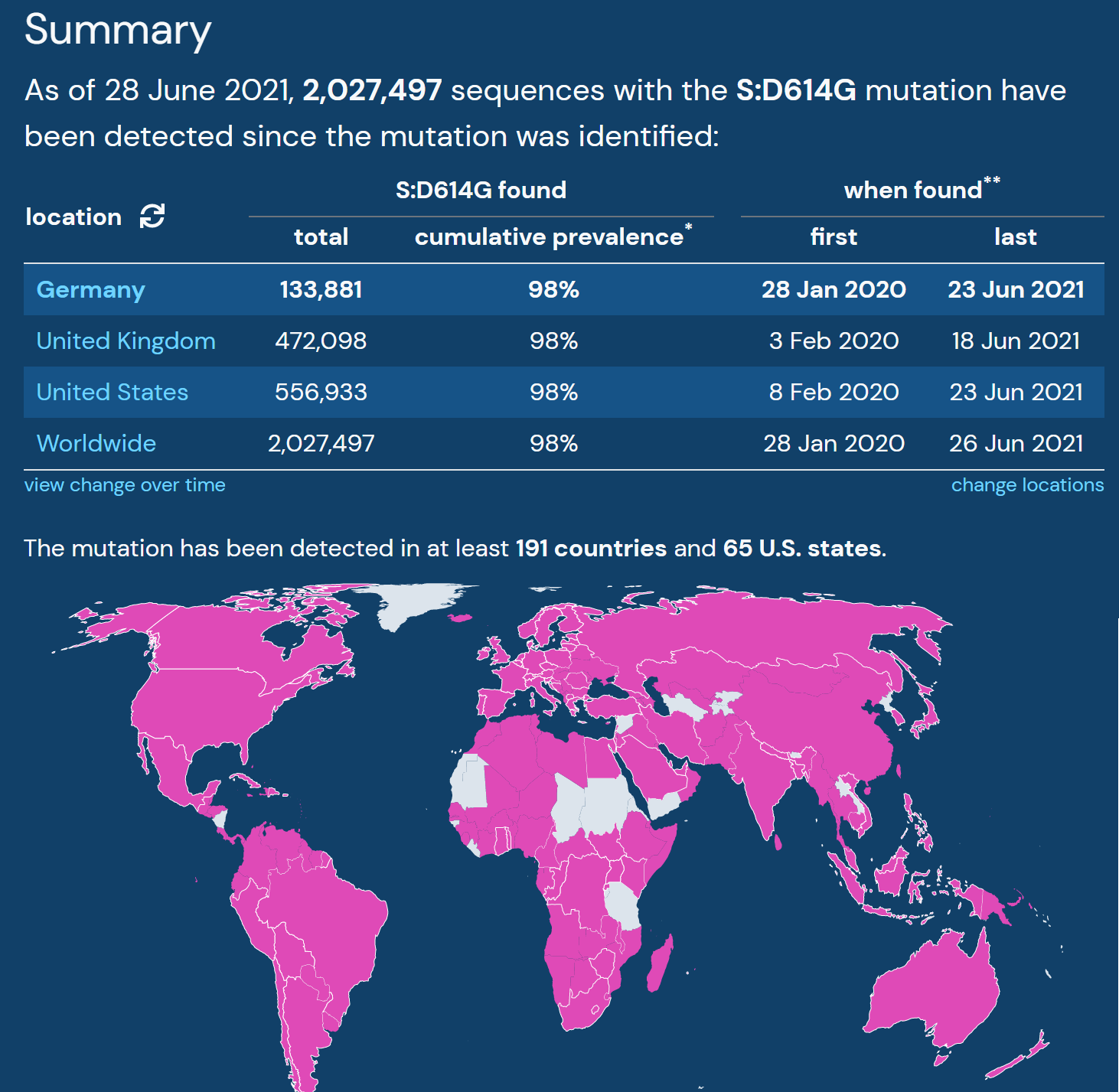 |
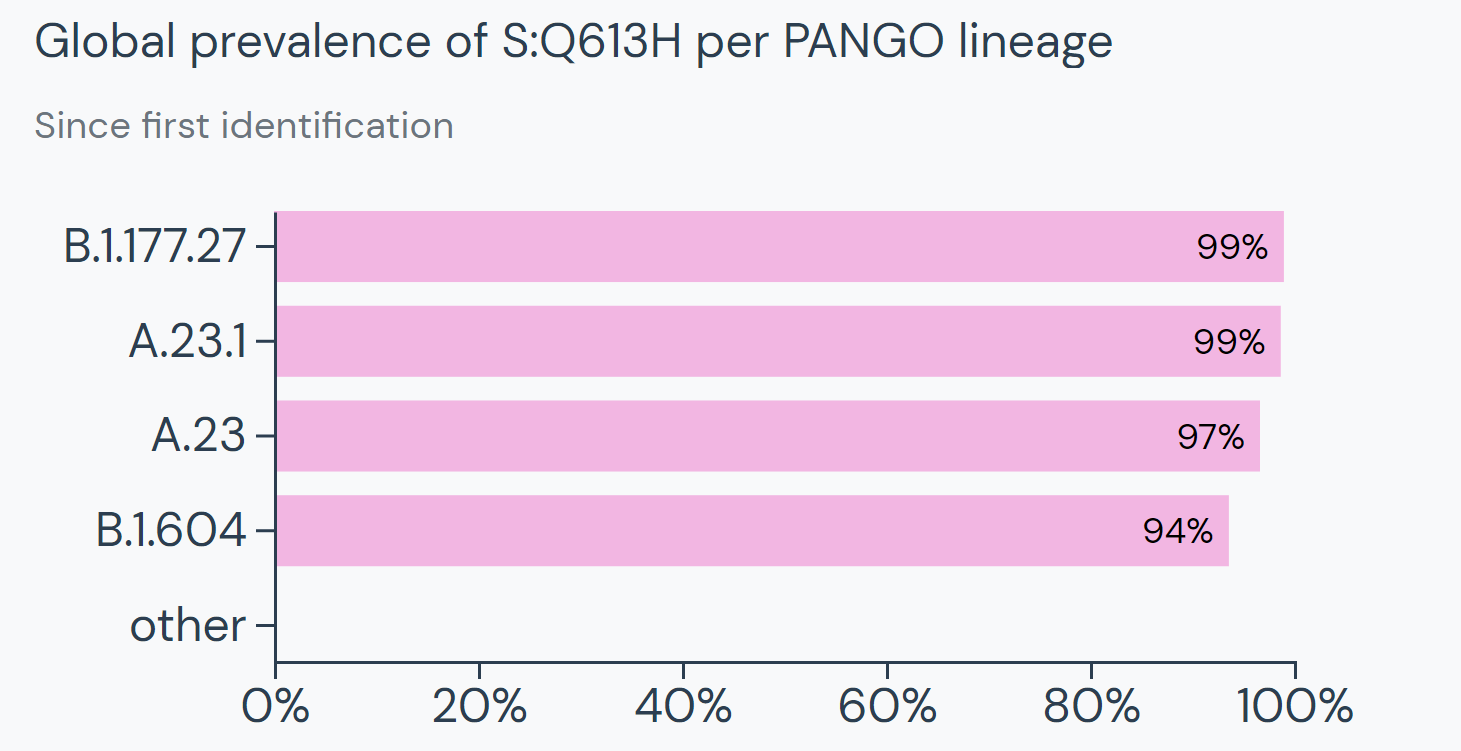 |
|---|---|
| S:Overview of S:D614G prevalence from outbreak.info | Lineages containing S:Q613H from outbreak.info |
Outbreak.info aids researchers in tracking the prevalence of mutations in SARS-CoV-2 lineages, and the prevalence of lineages in countries around the world. From breaking down and visualizing lineages by the mutations that define them, to aggregating global prevalence data by location, the scientific community now has free access to a key resource. Thanks to worldwide data-sharing enabled by GISAID, Outbreak.info is able to support the open-source line-of-attack driving success in the fight against the pandemic.
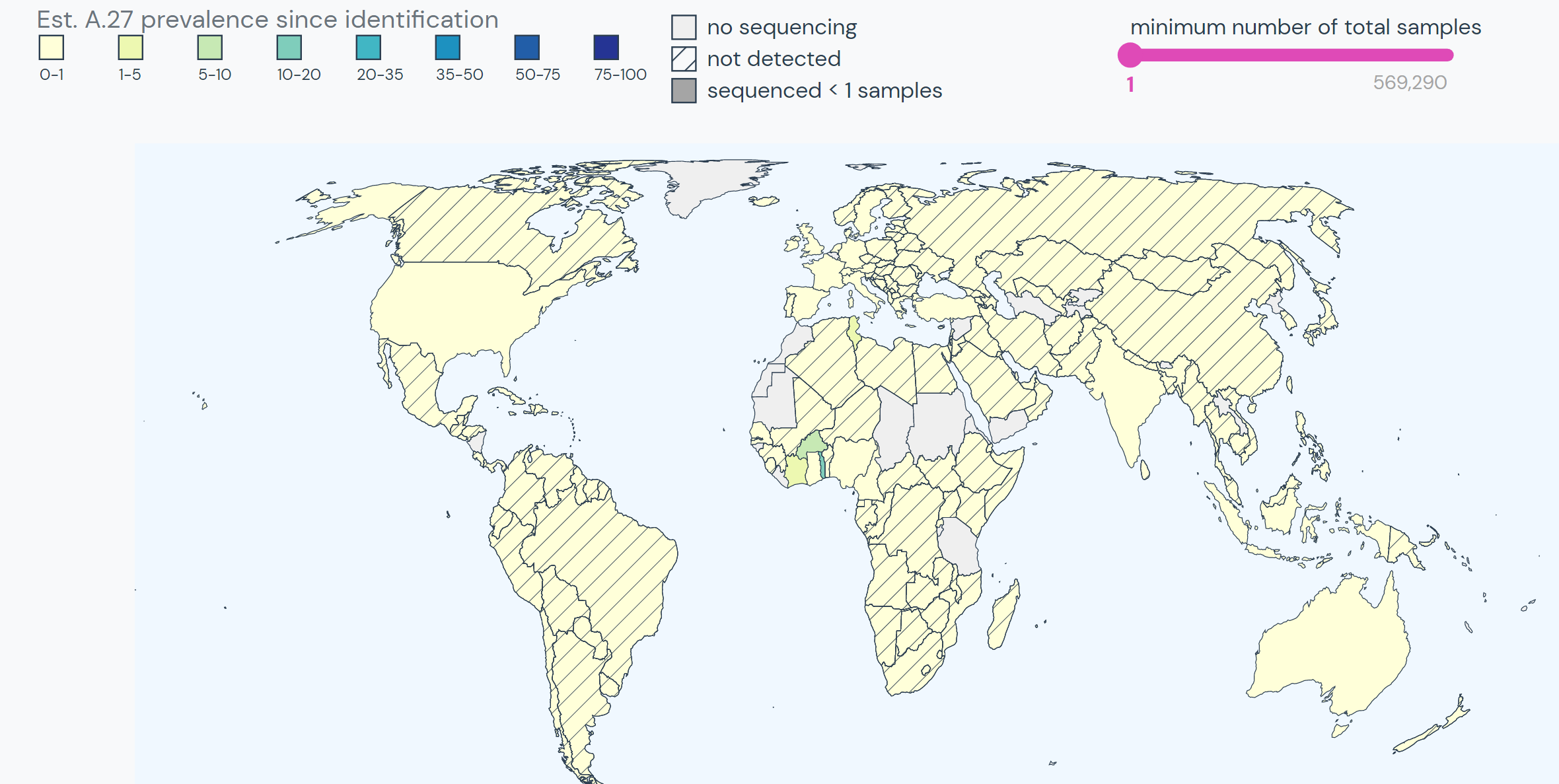
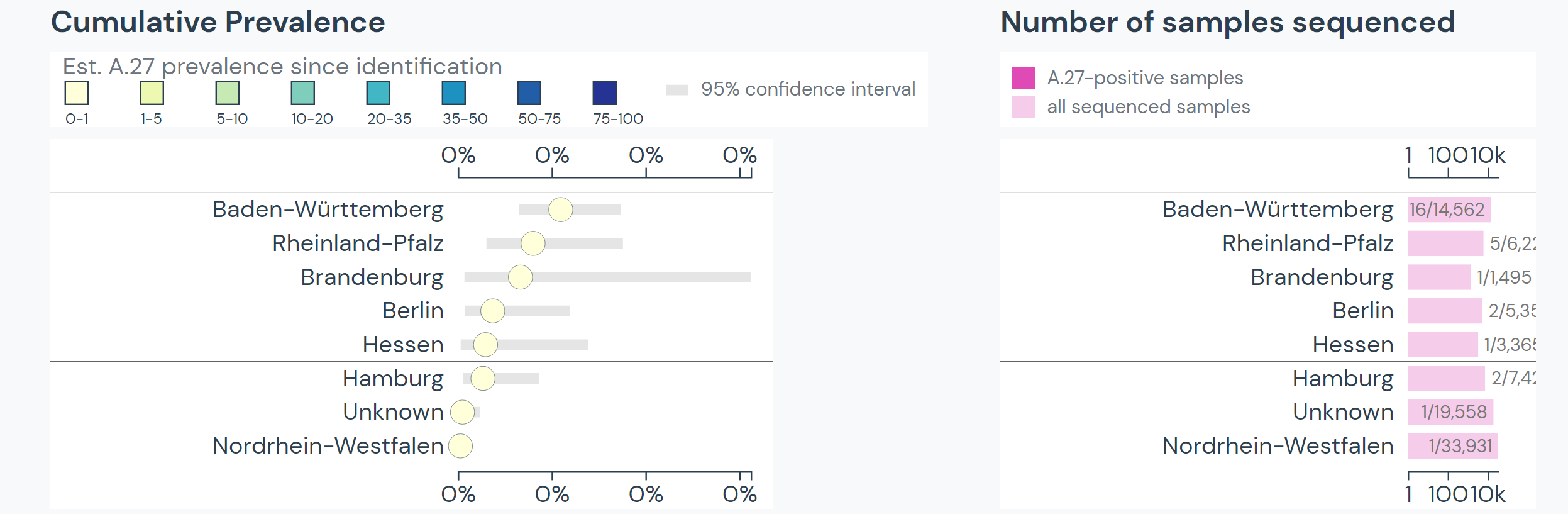
Calvignac-Spencer, S., Hölzer, M., et al. published their research in the Virological article, “Emergence of SARS-CoV-2 lineage A.27 in Germany, expressing viral spike proteins with several amino acid replacements of interest, including L18F, L452R, and N501Y in the absence of D614G.”
For more Outbreak.info stories, click here.