The emergence of COVID-19 was met with an unprecedented level of worldwide scientific collaboration, including the sequencing of over 10 million SARS-CoV-2 genomes shared via GISAID, making it the most sequenced virus in history. To make this sequencing data more accessible and understandable, Outbreak.info created interactive tools to track emerging variants based on publicly available data provided by GISAID. In collaboration with GISAID, Outbreak.info is proud to announce the release of an R package that enables near real-time analyses of all underlying genomic data on outbreak.info shared via GISAID. Through the R package, users can access the data visualized by outbreak.info’s Lineage|Mutation Tracker, Location Tracker, and Lineage Comparison tool.
Outbreak.info’s genomic tools are updated daily with GISAID data, providing free access to a database of over 10 million SARS-CoV-2 sequences. An average of 10,000-15,000 new sequences are added each day. Outbreak.info visualizes many key data variables for tracking the evolution and spread of Variants of Concern (VOCs). This makes it easier for researchers to use the data collected by GISAID, allowing them to focus on data interpretation and eliminating duplicative efforts. The R package allows researchers to download all of the raw data to support their own investigations or combine with other information for downstream analyses and visualizations.
This R package harnesses the power of BioThings Suite, developed by the Wu and Su labs at Scripps Research, which allows researchers to rapidly create high performance and easily scalable APIs that provide access to biomedical data. Genomic data is processed using the Bjorn pipeline, developed by the Andersen lab at Scripps Research, which relies on minimap and gofasta.
Outbreak.info also enables access to COVID-19 epidemiology data through the Epidemiology API and its library of aggregated COVID-19 journal articles, preprints, clinical trials, and datasets through the Resources API, as well as through the R package.
Outbreak.info supports open science and FAIR data practices by making all data and tools on the site publicly available. Outbreak.info and GISAID believe open data solutions are key to solving the pandemic and hope this collaboration will aid researchers in unearthing more about the evolution of SARS-CoV-2. This API serves to advance the interoperability of genomic data submitted by thousands of institutions and individuals around the world.
Lineage|Mutation Tracking
Outbreak.info’s Variant Tracker offers insight into how combinations of lineages and mutations are changing over time. The R package allows researchers to query the raw data to find the most prevalent mutations in a lineage, compare mutations across sublineages, discover the frequency of mutation combinations appearing in lineages, get lineage prevalence for locations, view prevalence changes over time, plot customized heat maps, and uncover what has been published about a variant in the literature. Read about these queries here.
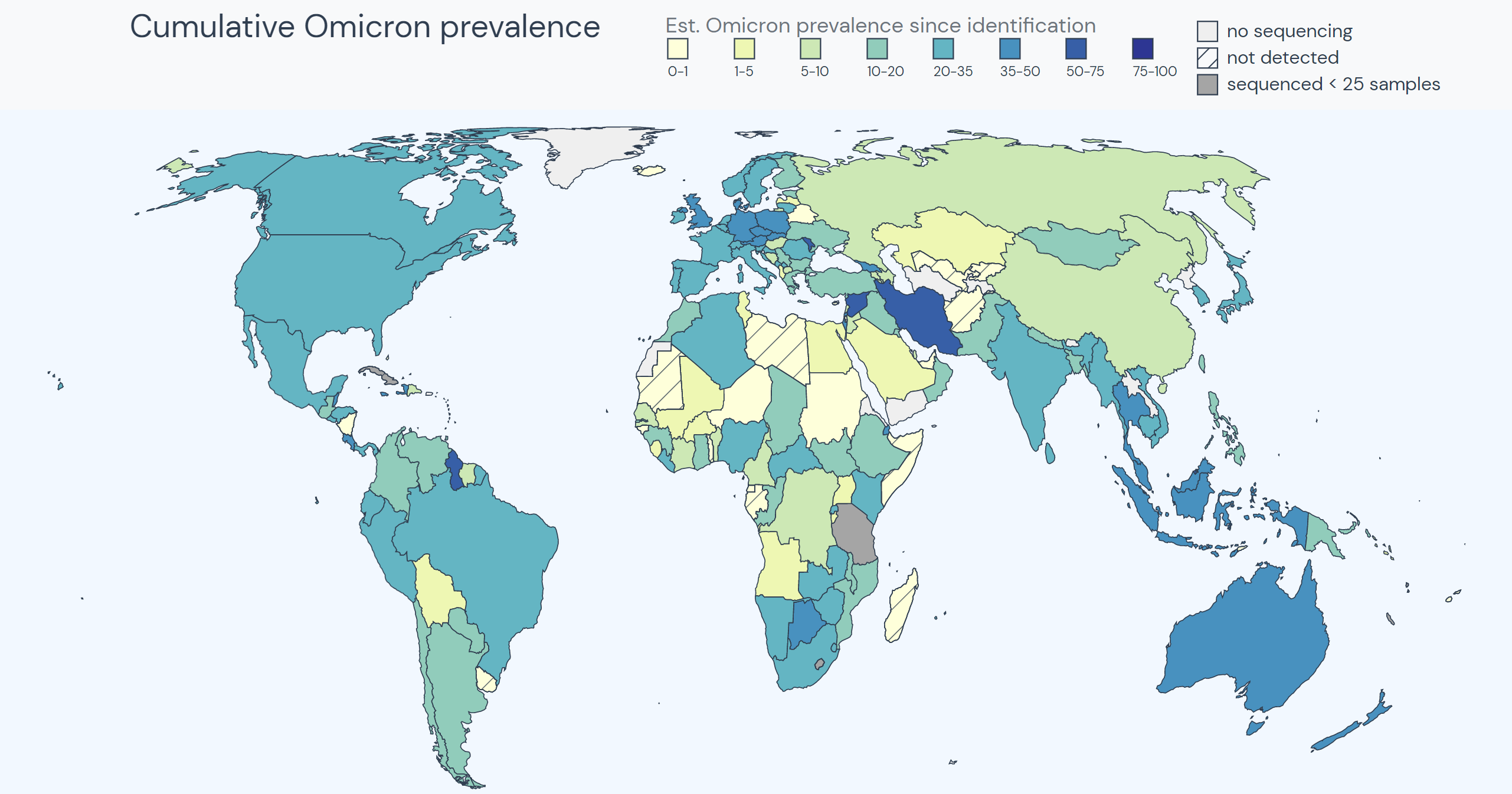
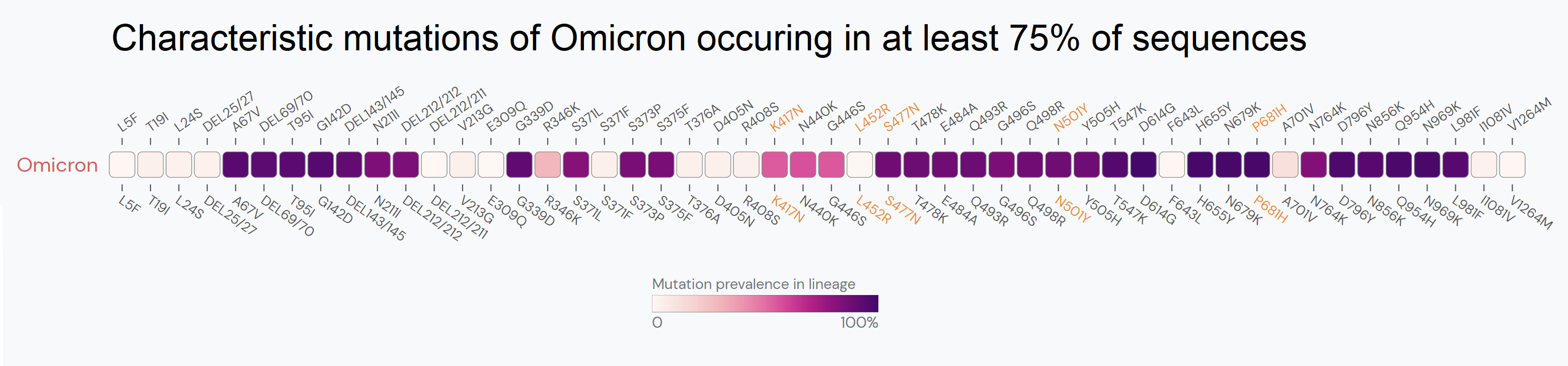
Location Tracking
Outbreak.info's Location Tracker provides investigation into the variants circulating in any country, state/province, or United States county. The R package enables researchers to compare variant prevalence within a location, view the prevalence of mutations or combinations of lineages and mutations per location, plot lineages in a location over time, find the Variants of Concern that are most prevalent in a location, or compare the prevalence of one variant over multiple locations. Read about these queries here.
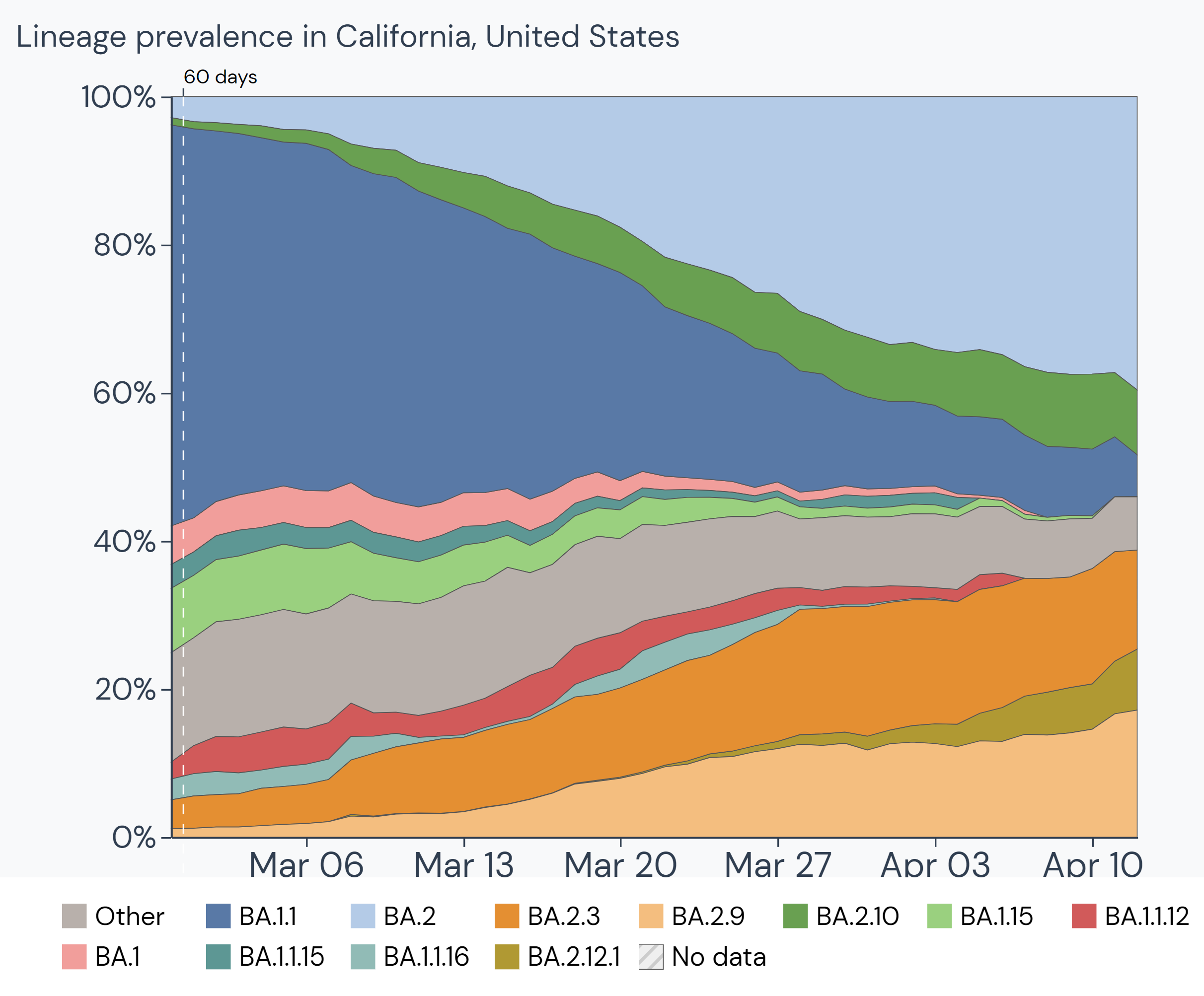
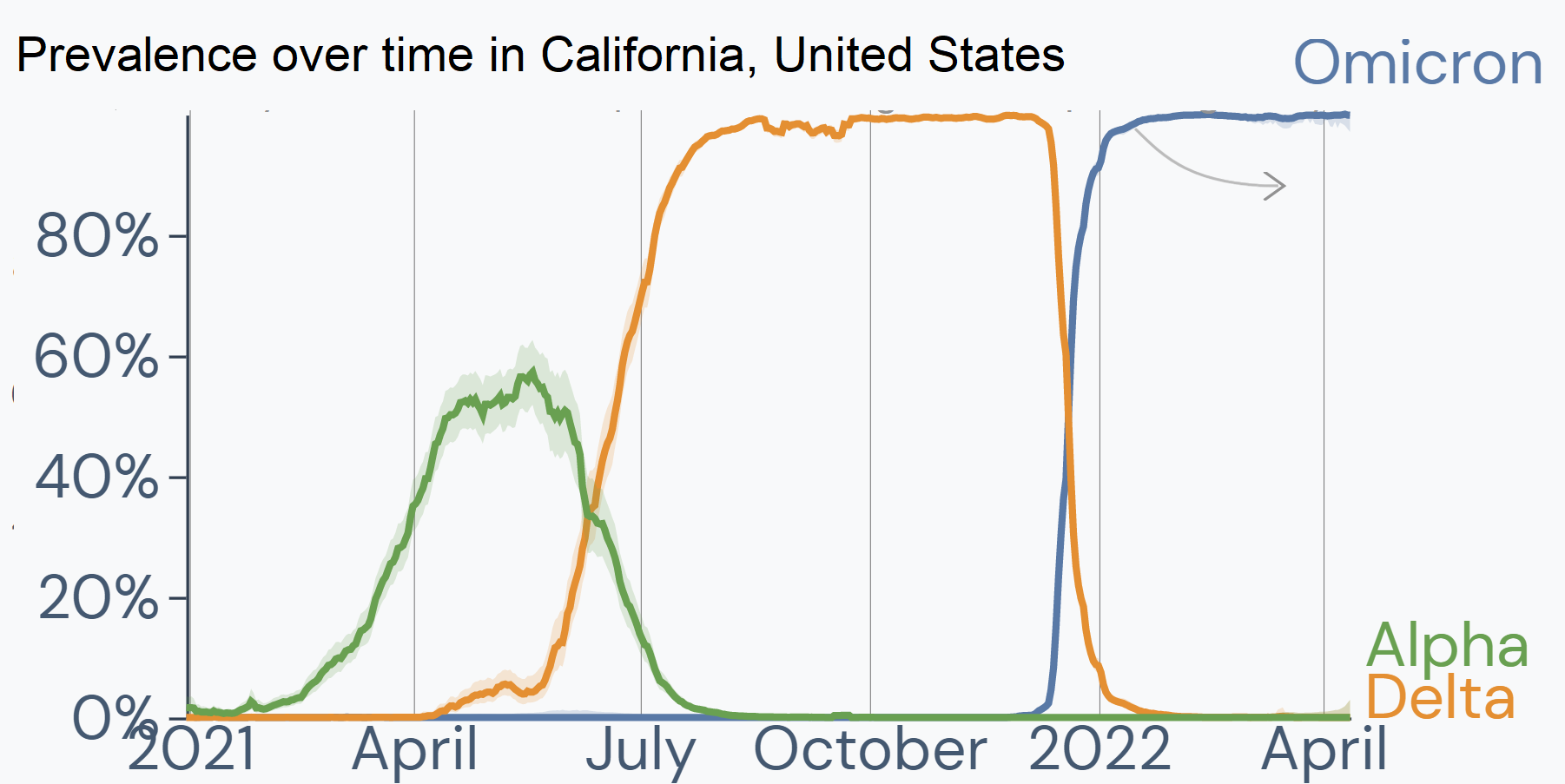
Getting started
- To protect data providers around the world, those accessing genomics data via the R package must be registered with GISAID (users can register here).
- Read about how to get started with the R package
- Access the Cases & Deaths and Research Library API directly
- Report any bugs or suggestions on GitHub